DNA sequence analysis is a fundamental technique in bioinformatics which enables scientists to study genetic material for various applications such as evolutionary studies, disease research, and genetic engineering. In this tutorial, we’ll guide you through building a simple DNA sequence analyzer using Python, following the approach outlined in the Caltech BiGE105 tutorial.
Prerequisites:
To follow this tutorial, you’ll need:
- Basic knowledge of Python programming
- Python installed on your computer (version 3.x recommended)
- Biopython library (install it using
pip install biopython)
Step 1: Reading a DNA Sequence
The first step is to load a DNA sequence from a file. DNA sequences are often stored in FASTA format, which consists of a description line(starting with >) followed by the sequence itself.
Code Implementation:
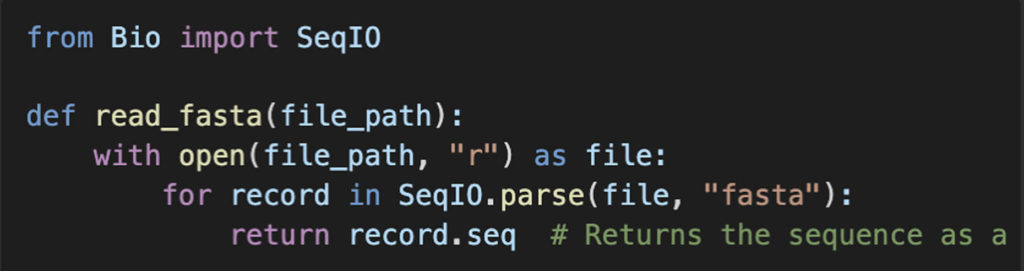
Explanation:
- We use
SeqIO.parse()from Biopython to read the FASTA file. - The function returns only the first sequence found in the file.
Step 2: Computing Basic Sequence Properties
Once we have our DNA sequences, we can compute some fundamental properties such as length, GC content, and nucleotide frequency.
Code Implementation:
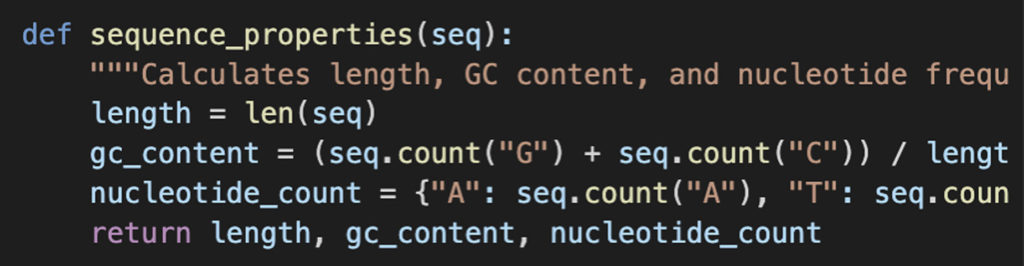
Explanation:
len(seq): Gets the total length of the sequence.seq.count("G") + seq.count("C"): Counts occurrences of G and C to determine GC content.seq.count("A"), seq.count("T"), ...: Computes individual nucleotide frequencies.
Step 3: Transcription (DNA to RNA)
Transcription is the process of converting a DNA sequence into an RNA sequence by replacing thymine(T) with uracil(U).
Code Implementation:

Explanation:
- The
replace("T", "U")function substitutes thymine with uracil.
Step 4: Finding Complementary and Reverse Complement Sequences
The complementary sequence replaces each nucleotide with its complementary base pair: A↔T, C↔G.
Code Implementation:

Explanation:
str.maketrans("ATGC", "TACG"): Creates a translation map to swap nucleotides.seq.translate(complement_map): Applies the translation to create the complement.[::-1]: Reverses the sequence to get the reverse complement.
Step 5: Finding Specific Motifs or Substrings
Biologists often search for specific motifs(subsequences) within DNA sequences.
Code Implementation:

Explanation:
seq.startswith(motif, i): Checks if the motif starts at positioni.[i+1 for i in range(len(seq))]: Collects all occurrences (1-based index).
Step 6: Running the DNA Analyzer
Now, let’s integrate everything into a script:
Code Implementation:

Explanation:
- Loads a DNA sequence from a FASTA file.
- Computes sequence properties like length, GC content, and nucleotide frequency.
- Converts DNA to RNA.
- Computes the complement and reverse complement.
- Searches for the motif
ATG. - Prints the results.
This simple DNA sequence analyzer allows you to load a sequence, calculate basic properties, transcribe it into RNA, find complementary sequences,s and locate motifs. With additional enhancements, you can expand this into a powerful bioinformatics tool. Try experimenting with real biological datasets that can be found on PubMed and extend functionality for applications like protein translation and mutation analysis.
Happy coding!
